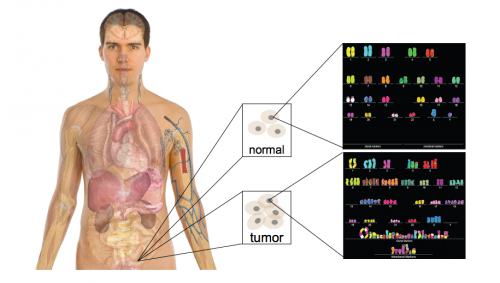
DNA, the key to our individuality and the unique traits that make us who we are, is also the key to many of our medical problems. However, due to technological limitations we have not been able to fully utilize the information stored in DNA until recently. Advancements in technology have made full genome sequencing faster and cheaper, making its information accessible to researchers across the nation. One such research team, Professor Mark Gerstein’s lab at Yale University, recently made a breakthrough by collaborating with groups that have extensive databases of human genomes. The joint teams discovered specific regions of the genome which act as signals for particular types of cancer. These regions are termed “cancer triggers.”

The Target of the Project
While the human genome was once completely unintelligible, in 2003 a partnership between the National Institutes of Health and the Department of Energy succeeded in fully sequencing the first human genome after thirteen years of research. Now, the human genome is no longer an end goal in genetic research, but a means to an end. The advent of new projects such as ENCODE, which aims to characterize functional elements of the genome, and TCGA, the cancer genome atlas, provide extensive databases of human genomes. These projects were also the resource for the raw data used by Gerstein and Dr. Ekta Khurana’s team to determine specific cancer trigger mutations in the human genome.

The human genome contains over three billion base pairs, less than one percent of which code for genes. Although 99 percent of the human genome is non-coding, these regions are still important. Non-coding regions actually form the primary focus of study for many bioinformatics projects, because gene regulation occurs within these segments. According to Gerstein, “The genes may be like light bulbs — where one can actually see the light — but the non-coding region is like the wire with all the essential controls and switches, and that is where the regulatory apparatus is located.” But how do you find the regions in non-coding segments of DNA that are pertinent to essential genes? The answer lies with evolution: The most important sections of DNA have been conserved in living organisms because their functions are vital for survival.
Out of the three billion base pairs, there are about 5,000 somatic variants that differ from one person to the next; however, not all 5,000 of them are important in gene regulation or determining disease. Historically, the important variations were identified as those within actual genes, but because some non-coding segments influence the regulation of genes, we now recognize the importance of investigating other somatic variations further. The first goal in this project was to find the somatic variants that overlap with the conserved and functional regions of the non-coding DNA, and then to mark those regions for further study.

The “Elaborate Filter” for the Triggers
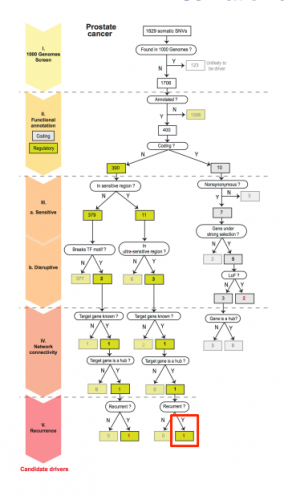
The sequencing and data collection for the project necessitated collaboration between many institutes and universities, but Khurana’s team lead the primary analysis. In order to narrow down the 5,000 variants to approximately ten important mutations, the researchers needed an elaborate filtration system. By cross-referencing thousands of human genomes from ENCODE, the truly conserved regions were first determined. Then these regions were ordered according to their functional annotation, specifically with regards to the importance of each region in gene regulation.
Finally, the 5,000 variations (excluding those within genes) were overlapped and ranked according to their occurrence in a prioritized region of the non-coding genome. Genomes from TCGA were then used to identify specific cancer variations that occur in conserved regions. Once the variants were narrowed down, they were ranked according to the ordering of functional annotations. Finally, the variants were prioritized by recurrence in the same region for multiple cancer genomes. Statistical analyses determined the identified cancer-driving mutations to be significant. By applying this filter and creating “decision trees” for over 90 prostate, breast, and medulloblastoma cancer genomes, Khurana and her team identified over 100 cancer-driving mutations.
Challenges in Identifying the Triggers
When someone mentions biology research, the first things that come to mind are usually Petri dishes and mice, but Khurana’s project was conducted on computers. While it may seem that technology would eliminate many process-related issues, the real challenge of this project did not lie in its execution, but rather in conceptualizing the biological framework of the problem as a measurable test. For such a bioinformatics project, this proved quite challenging. There are over three billion base pairs in the genome, and this study utilized thousands of genomes, which involved trillions of data points. Experimental testing necessitates careful accounting of technical artifacts, and with thousands of genomes the required controls can become complicated. The grand scale at which it is now possible to analyze biological systems means that scientists are now limited less by the technology and more by the boundaries of their own creativity.
Triggers as Potential “Targets”
The scientific world has known of the genome’s power since Watson and Crick described the double-helix model for DNA in 1953; however, the vast amount of information stored in the genome has been largely inaccessible until recent years. With the invention of better DNA sequencing machines as well as improved computer power, the determination and analysis of the genome was made possible, and since 2003 the application of scientific investigation upon that information has led to the discovery of almost 2,000 genes linked to diseases. Gerstein’s lab and collaborators have ventured into a relatively unmapped region of DNA: the non-coding area. If the past is any indicator, further research in these regions will provide rapidly increasing insight to our inner genetic workings and reveal more targets to control disease. This path of progress regarding insights into the genome is a common theme in science. First, the sequencing of the genome was the target; then it became the tool or the “trigger” for new discoveries. These new discoveries can be specifically “targeted” in further investigations.

The identified triggers are now potential candidates for study by cancer biologists. Results of mass scale experiments tend to provide direction for many in-depth experiments, as is the case here. “Most people don’t know what to do with noncoding variants,” said project leader Khurana, “Our tool can be used to prioritize these variants for further follow up.” Indeed, the higher priority trigger points will be extensively characterized to confirm that they are, in fact, cancer drivers and to further examine how exactly each one is involved in the development of a particular cancer. Only then does the possibility of personalized therapeutic approaches come into consideration. Moreover, this approach of determining cancer triggers can be applied more generally to determine countless other disease triggers, truly setting the foundation for further progress in fighting disease.
About the Author:
Deeksha Deep is a sophomore Molecular Biophysics & Biochemistry major in Morse College. She is on the business team for the Yale Scientific Magazine and the beat editor for the Yale Journal of Public Health. She works in Professor Spiegel’s lab studying cell surface reconstruction in bacteria and vaccine design.
Acknowledgements:
The author would like to thank Professor Mark Gerstein and Dr. Ekta Khurana for their time and enthusiasm about their research.
Further Readings:
- Khurana, E. et. al, “Integrative Annotation of Variants from 1092 Humans: Application to Cancer Genomics,” Science 342 (2013), DOI: 10.1126/science.1235587
- The 1000 Genomes Project Consortium, “A map of human genome variation from population-scale sequencing,” Nature 467 (2010), 1061-73.
