Professor Patrick Loria has always been fascinated by the fundamentals — internal motions in proteins and the functions of those proteins in the body. Enzymes, the proteins that work to catalyze reactions, are certainly not static. The Loria lab investigates the internal motions of biological enzymes, an area of study that is crucial to drug discovery efforts. Using nuclear magnetic resonance spectroscopy of proteins (protein NMR), the Loria lab studied how phosphate, sulphate, and acetate buffers can influence motions and illuminate the functions and allosteric regulation of enzymes.
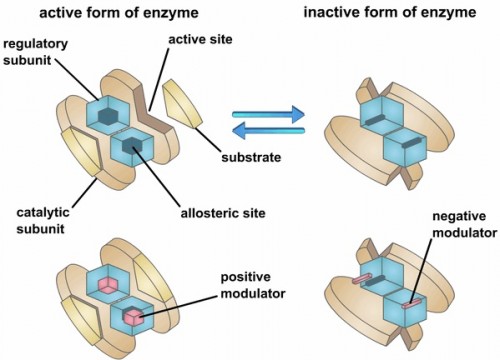
Allosteric enzymes are proteins that change their conformation when bound to an effector protein. This change in conformation can either activate or inactivate the enzyme. Although functional allosteric enzymes regulate many classic pathways, the biochemical mechanism of action of most of these enzymes is still very unclear. The Loria lab works in collaboration with Yale Chemistry Professor Victor Batista, who is an expert in computational chemistry, to understand and rationally alter allosteric enzymes.
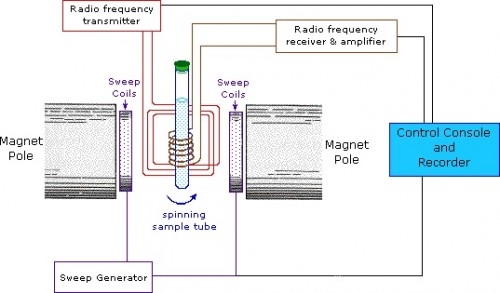
Techniques and Tools of the Loria Lab — A Quick Primer
The Loria lab used NMR spectroscopy extensively to analyze the influence of buffers on enzyme motions. In NMR, the nuclei of atoms, depending on whether they possess an intrinsic form of angular momentum (spin property), behave like small magnets. Upon immersion of these atoms into a static magnetic field of the NMR spectrometer while maintaining exposure to a secondary oscillating magnetic field, an NMR signal is produced. This is a physical phenomenon in which nuclei in a magnetic field absorb and re-emit electromagnetic radiation at a specific resonance frequency. The chemical shift of a nucleus is the difference between the resonance frequency of the nucleus and a standard reference compound (e.g. tetramethylsilane, which is assigned a chemical shift of zero). This quantity is reported in parts per million (ppm) and given the symbol delta, Δ. The chemical shift differs for the various amino acids (residues) of an enzyme. The protein NMR signal helps scientists classify the protein and better understand its dynamic properties.
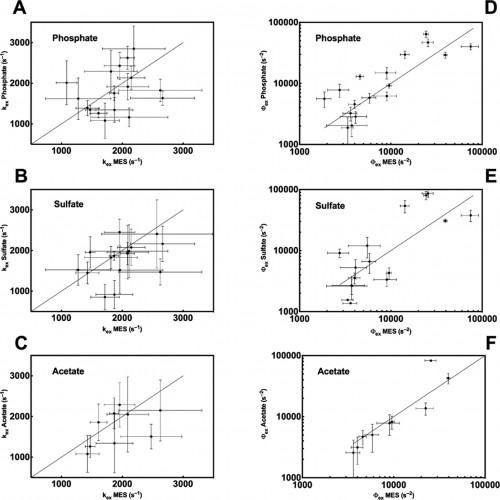
The Loria lab measures spin-spin relaxation times using relaxation dispersion experiments, which are pulse sequences applied during spectroscopy. The term relaxation describes several processes by which nuclear magnetization prepared in a non-equilibrium state returns to the equilibrium distribution. In other words, relaxation describes how fast spins “forget” the direction in which they are oriented. Spin-spin relaxation is the mechanism by which the transverse component of the nuclear spin magnetization vector exponentially decays towards its equilibrium value of zero. The metric (Rex), which is commonly used to analyze results from the relaxation dispersion experiments, depends on conformational exchange parameters ϕex and kex. The parameter ϕex takes into account the equilibrium population of a pair of inter-converting conformers as well as the difference in chemical shift between the two conformations. The parameter kex is the sum of the forward and reverse rate constant of the conformational states.
Investigations at the Loria Lab
Imidazole glycerol phosphate synthase (HisF), an allosteric enzyme needed for the survival of plant pathogens and some pathogenic bacteria, has been studied extensively in the Loria lab. If the mechanistic properties of this enzyme are better understood, scientists could use small molecules to inhibit the enzyme and eventually provide a therapy for those affected by the pathogen. Using protein NMR on HisF, the team noticed chemical shifts that changed based on the buffer being used. When this phenomenon was explored in the literature, the lab found research on the pH dependence of atomic resolution structures that complemented the solution NMR experiments but found very little work that investigated how solute and buffer molecules interact to change enzyme conformation. Therefore, the lab embarked on a set of control experiments to illustrate the importance of choosing the most appropriate buffer as common solute molecules can alter observed enzyme millisecond motion.
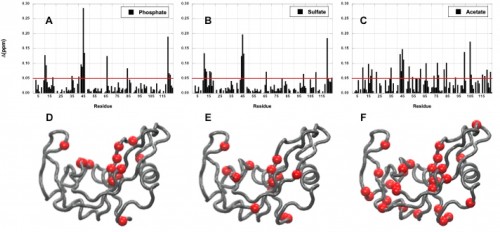
In a study led by Madeline Wong ’13, the Loria lab implemented solution NMR to examine the effects of different functional groups in the enzymes HisF, ribonuclease A (RNase A) and triosephosphate isomerase (TIM). Phosphate, sulfate, and acetate were used as buffers as these functional groups have shown a number of ways to interact with proteins of interest. The effects of these buffers on the chemical shift perturbation and millisecond conformational exchange motions of the enzymes were compared to the effects of reference buffer systems HEPES and MES. When the NMR experiments were performed to examine chemical shifts and dynamics of the three enzymes, the scientists kept the pH, ionic strength, and temperature constant so that the buffer was the only independent variable. The researchers subsequently performed relaxation dispersion experiments, which showed significant solute-dependent changes for all of the enzymes. Lastly, when the X-ray crystal structure for all three enzymes was analyzed, there was a significant presence of phosphate ions at the enzymes’ active sites, clearly illustrating that an anionic buffer could alter conformational dynamics of enzymes. In future research, the lab will perform enzyme assays with and without different buffer components to see if buffers can inhibit enzyme function.
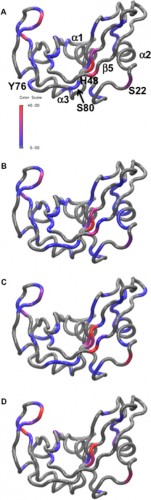
Through the results of this study, the Loria lab has illustrated the importance of buffers, and how buffers can confound biochemical experiments. “Typically buffers for biochemical studies are chosen as a matter of experimental convenience. What Madeline’s experiments show is that buffers can play a more active role, often altering the function and dynamics of the system under study,” stated Loria. Although addressing this issue in the lab may be difficult, as even the best buffers could affect enzymatic conformation, these results have highlighted an important procedural issue.
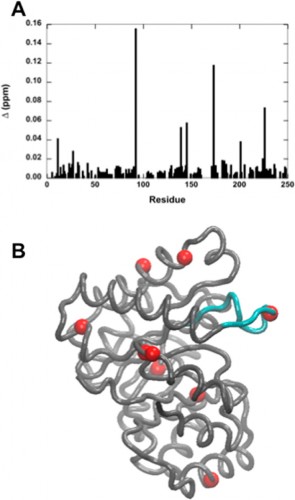
On a local scale, the results of Wong’s study continue to inspire current Yale students. Tim Caradonna ’15 is now starting his second year in the Loria lab. When asked what drives scientists such as himself in this type of basic research, Caradonna stated, “The goal of basic research is to solve fundamental problems, which can provide an established context for more applied research in the future. Experiments such as Madeline’s are important controls; if the buffer has changed the dynamics of a protein, you’ve potentially affected its function or activity.” On a larger scale, knowledge of how buffers can affect enzyme activity offers important insight into the future of drug development and other fields of biochemical experimentation.

Courtesy of Patrick Loria.
About the Author: Jayanth Krishnan is a junior Molecular, Cellular & Developmental Biology major in Morse College. He is the Journalism Chair for Synapse – Yale Scientific Magazine Outreach and works as a remote affiliate at Professor Kellis’s lab at MIT.
Acknowledgements: The author would like to thank Professor Loria for his time and for his enthusiasm about his research.
