It is not often that a pathogen is compared to a “crafty burglar,” yet the description is apt for the bacteria that causes Legionnaire’s disease, a severe form of pneumonia. When Legionella pneumophila enters the body, it is ingested by macrophages and stored in large cellular cavities known as vacuoles .
Normally, the vacuoles would then fuse with lysosomes, and bacteria-degrading enzymes would destroy the pathogen. However, L . pneumophila alters the transportation system of the vacuole so that it instead fuses with the endoplasmic reticulum (ER), a nutrient-rich environment for replication.
In addition to infecting humans, this bacteria also infects amoebae, single-celled eukaryotes; it is this unique ability to cross such a wide evolutionary gap that piqued the interest of Craig Roy, associate professor of microbial pathogenesis. “L.pneumophila learned to replicate in amoebae. It never coevolved with mammals, so its ability to replicate in humans suggested to us that it must be targeting highly conserved proteins,” Roy said.
Roy aims to understand how bacteria have become pathogenic. He chose L.pneumophila as a model organism, because, despite a major outbreak in 1976, the bacteria is not contagious. In fact, L. pneumophila can be found in any given sample from a freshwater body.
As its name suggests, it can cause severe pneumonia if inhaled in extremely high numbers, although early treatment with antibiotics normally ends the infection. Roy believes that understanding its mechanism of evading the immune response may help combat other more debilitating pathogens, such as Myobacterium tuberculosisand Chlamydia. These pathogens use similar mechanisms, although they are coupled with additional evasion techniques due to their coevolution with humans.
In a recently published paper in Nature, Roy and colleagues elaborate on the genetic basis of L. pneumophila’s success. By infecting mice macrophages with the bacteria, the Roy lab was able to illuminate the details of the infection mechanism.
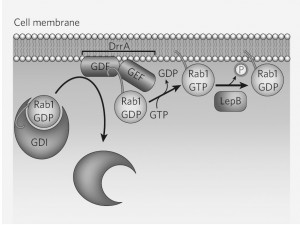
L. pneumophila targets a highly conserved protein known as Rab1, which regulates the transport of ER vesicles in macrophages. Guanine nucleotide-exchange factor (GEF) is a protein that activates Rab1. By isolating this protein in L.pneumophila mutants that cannot recruit Rab1, Roy and colleagues discovered a bacterial protein that acts as GEF. They called it DrrA, or “defect in Rab1 recruitment A.”
DrrA also acts to displace a dissociation inhibitor in the first step towards activating Rab1. Dr. Roy and his colleagues also found another bacterial protein,Legionella effector protein B (LepB) that inactivates Rab1 after it has altered theLegionella-containing vacuole (LCV). LepB mimics the macrophages GTPase-activating protein (GAP), which converts the active Rab-GTP into the inactive Rab-GDP.
Since 1992, Roy has been working to understand the molecular and cellular events that enable microbes to evade host immune systems. Back then, when almost nothing was known, he would begin by generating random mutants, identifying the critical genes, and finally understanding the proteins that the genes encode. Today, he and his lab have a much better understanding of the genes and proteins that enable L. pneumophila/ to replicate in amoebae and macrophages.