
Human health and disease are intimately connected to the bacteria around us. Throughout history, bacteria have afflicted the human race with infections ranging from minor food poisoning to massive epidemics of cholera and bubonic plague. On the other hand, a whole host of beneficial bacteria are carried around in the human gastrointestinal tract, helping us to thrive. With such diverse biological effects, bacteria hold the key to uncovering the drugs of tomorrow.
The Crawford Laboratory, located on Yale’s West Campus, is especially aware of bacterial diversity and the potential biomedical relevance of how bacteria interact with their hosts. The lab’s principal investigator, Jason Crawford, strategically mines bacterial genomes in search of the biologically active small molecules these microorganisms use to fulfill their unique roles throughout nature. To this end, the lab takes advantage of the readily accessible genomic data available through modern sequencing technology and locates orphan gene clusters — those without any assigned function. Such gene clusters are often acquired in the gain of an evolutionarily advantageous trait, making them ripe for the discovery of novel bioactive compounds.

Beginnings of the search
Though Crawford’s genome screens and orphan gene clusters are new, the search for medicines from bacteria is not. Ever since Alexander Fleming’s isolation of penicillin from a fungus in 1928, scientists have been intrigued by natural treatment products derived from microorganisms. Investigation of the bacteria behind diseases like cholera has revealed that some pathogenic and mutualistic bacteria interact with humans and other host organisms using secondary metabolites, small molecules that have evolutionary purposes beyond normal growth and reproduction. Bacteria produce these metabolites as a way of surviving within a host and use them to affect cellular machinery, elude the immune system, or outcompete other bacteria.
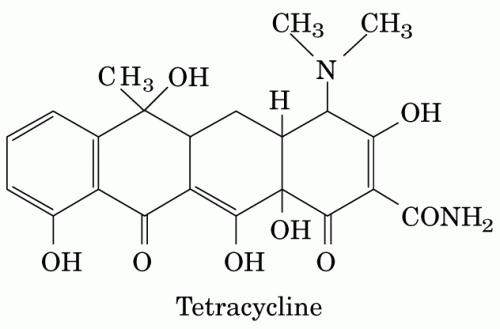
The ability of such compounds to target proteins with high specificity has made natural products ubiquitous in drug discovery and pharmaceutical development. According to David Newman at the National Cancer Institute, just over half of the small molecule drugs today are natural products or natural product-derived. Several antibiotics, including the widely used tetracycline class first isolated from Actinobacteria, are derived from natural products that exist as secondary metabolites in bacteria. In addition, natural product discovery in plants and fungi has led to drugs ranging from Fleming’s penicillin to the anticancer agent Taxol.
The advantage of natural products over human-designed drugs lies in their evolutionary optimization. Designed drugs may result in unintended, deleterious interactions with other body systems or proteins that outweigh their benefits. “If you want to design a molecule, there’s no evolutionary selection in that process,” said Crawford, “which means that the chances of a molecule from a synthetic library having a pharmacologically relevant or functional role is actually quite low.” Natural products, on the other hand, have been sculpted by evolution to affect specific proteins or processes in hosts or competing microorganisms.
Mining for natural products
Crawford and other researchers who seek to isolate the products of evolution have long realized their potential. However, the high costs associated with genomic sequencing just five years ago and the relative lack of sequence information at that time traditionally limited research like Crawford’s. The search for natural products has instead relied more commonly upon finding novel bacterial and fungal species in soil and vegetation samples from around the world. Extracts from these species can be screened for therapeutic or anti-bacterial functions in eukaryotic and other bacterial cells.
“Genomics-guided approaches are newer,” said Crawford. “The whole field is only about 10 years old, but the explosion in genomic sequencing over the past five years is what has really catapulted it forward.” Now, comparing bacterial genomes before and after the acquisition of new traits allows Crawford to identify gene clusters that encode small molecules responsible for what he calls “evolutionary leaps.” For example, a bacterium that makes the leap from the insect gut to infecting humans must gain new immunosuppressant or antibacterial traits in order to survive and compete. If these traits can be pinned to an orphan gene cluster acquired in the leap, it’s likely that the gene cluster codes for useful metabolites with these exact functions. The same compound the bacterium uses to fight off other pathogenic bacteria can be isolated and developed into a novel, potent antibiotic.
The orphan clusters that Crawford identifies do not necessarily code directly for small molecules. Bacteria often make use of enzymes like polyketide synthases and nonribosomal peptide synthetases, complexes of several proteins that piece together natural products in assembly line sequences. Tetracyclines and other antibiotics, immunosuppressants, and anticancer drugs are all synthesized by these enzymes. Other biocatalysts and machinery for exporting or processing small molecules could also be found nearby in gene clusters. “There was a point in time long ago when nobody knew what a polyketide synthase was,” Crawford explains. “So the question is: what other enzyme systems are left that we haven’t yet discovered?”
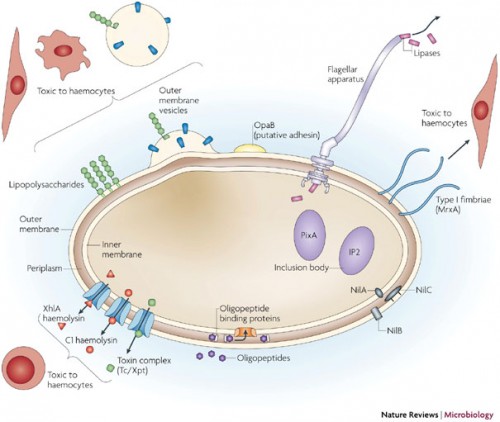
In a recently published review, Crawford discussed one system in particular he has worked with, in which such enzymes are coming to light. His work involves two bacteria, Xenorhabdus and Photorhabdus, found in the gut of nematodes. The nematodes, which act as insect parasites, have a developmental response that causes them to regurgitate the bacteria when inside a host. The bacteria respond by producing insecticides to kill the insect and antibiotics to outcompete competitors. These responses are exactly what Crawford looks to in his search for natural products. “A developmental biologist would say the developmental response is regurgitation and then look for the genes that cause this response, but a chemist focuses on the small molecules controlling this response,” he said.

Not only do the bacteria produce molecules capable of inducing regurgitation in nematodes, they must also work to suppress the insect’s immune system, kill the insect, and then outcompete other bacteria or fungi. “Just there, there are about five biomedically relevant objectives in this simple life cycle: developmental modulators, immunomodulators, insecticides, antibacterials and antifungals,” said Crawford. Even the most basic bacteria-host interactions are rich sources of potential drugs, making research like Crawford’s particularly exciting for the pharmaceutical industry.
Looking to the future
Though they are still somewhat new at Yale, the Crawford group has several projects in the works. Studying bacteria like Xenorhabdus or Photorhabdus, they are focusing on identifying enzymes in gene clusters that can catalyze the synthesis of novel small molecule structures. “If you find new structural features the chance of finding new function is also higher,” Crawford explains. “Our version of a grand slam is identifying new biocatalysts that are involved in the synthesis of new structural features that then lead to new biology through the functions they encode.” This “grand slam” may not be so far off. Members have already functionally identified an enzyme in Photorhabdus with no known homologues and a separate novel small molecule class characterized by an unusual carbon-nitrogen bond linkage. In addition, the lab recently received a grant to investigate and isolate small molecules involved in bacterial ability to both suppress inflammatory bowel diseases and drive colorectal cancer in humans.
Given the incredible diversity of bacterial systems in nature, the possibilities for research seem endless. Whether applied to colorectal cancer in humans or the modulation of the immune system by nematode bacteria, the Crawford lab’s gene-to-molecule approaches have great potential for natural product discovery. A future in which genomic screening allows for the development of potent anticancer agents from even the most unlikely, gut-residing bacteria may be closer than we think. “A lot of researchers want to be a part of an already-developed history and fine-tune some of the pages in that history,” Crawford says. “We would like to see if we can start our own little chapter.”
About the Author:
Jake Allen is a junior Molecular, Cellular and Developmental Biology major in Ezra Stiles College. His past research has focused on the pathogenesis of Alzheimer’s disease and the effects of ketamine on signal transduction in depression.
Acknowledgements:
The author would like to thank Professor Jason Crawford for his time and guidance and for his work bridging the gap between chemistry and biology.
Extra Reading:
Maria I. Vizcaino, Xun Guo, Jason M. Crawford. 2013. “Merging chemical ecology with bacterial genome mining for secondary metabolite discovery.” Journal of Industrial Microbiology and Biotechnology.
David J. Newman. 2008. “Natural Products as Leads to Potential Drugs: An Old Process or the New Hope for Drug Discovery?” Journal of Medicinal Chemistry (51): 2589-2599.
Heidi Goodrich-Blair, David J. Clarke. 2007. “Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: two roads to the same destination.” Molecular Microbiology (64): 260-280.
Harald Gross. 2007. “Strategies to unravel the function of orphan biosynthesis pathways: recent examples and future prospects.” Applied Microbiology and Biotechnology (2): 267-277.
