The creation of induced pluripotent stem cells (iPSCs) is “one of the greatest things that has happened to biology in the last 20 to 30 years,” Andrew Xiao at the Yale Stem Cell Center enthused. And with good reason. iPSCs hold all the regenerative power of the embryonic stem cells (ESCs) they have been reprogrammed to mimic, while steering clear of the thorny ethical issues associated with the latter.
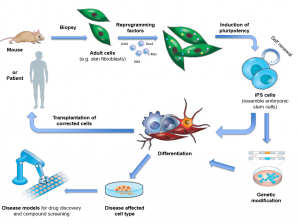
Image Courtesy of EuroStemCell and Michael Rossbach
Now, Xiao and his team of researchers have uncovered a powerful marker that can be used to distinguish the quality of iPSCs in mice, paving the way for advances that will allow these cells to revolutionize the fields of regenerative medicine, cell therapy, drug screening and disease modelling.
Not all iPSCs are created equal. While some indeed revert to a pluripotent state, ready to differentiate into any tissue in the body, the majority suffer defects in the reprogramming process, giving rise instead to unusable tissue or, more worryingly, to tumors. Thus, a prevailing question has been whether scientists can efficiently tell them apart.
The key may lie in the histone variant H2A.X. Scientists increasingly recognize that the histone complexes around which our DNA is wrapped play an important role in gene expression. They also know that while these complexes in general incorporate histone H2A, in a very small portion of our genome, H2A.X — a variant with a slightly different amino acid sequence — is deposited instead. Until now, the significance of this phenomenon was largely unknown.
Xiao and his team discovered that ESCs and high quality iPSCs share a characteristic pattern of H2A.X deposition across the genome, in marked contrast to defective iPSCs. Even more exciting was their finding that knocking out H2A.X in ESCs compromises their pluripotency, thus indicating that this variant is more than a marker: it plays a critical role in maintaining the developmental potential of ESCs and iPSCs.
How is it that a single protein variant exerts such great influence and predictive power over a complex a phenomenon as pluripotency? Lead author Wu Tao describes the immense frustration in trying to make sense of the mountain of data they generated — all 100GB worth. Even powerful bioinformatics tools initially did not yield any explanation for the pattern of H2A.X deposition they observed. Then, they realized that H2A.X deposition correlated with genes regulated by three differentiation factors, suggesting a mechanism of action: H2A.X remodels the chromatin in regions where it is deposited, thus regulating the expression of these critical genes.
Work remains to be done. H2A.X assays already hold important advantages over existing tests of stem cell quality, but the team envisions simplifying them further. They are also moving on to understanding why defective H2A.X deposition occurs and how it upsets the reprogramming process.

Xiao acknowledges that understanding the basic mechanisms is a tedious process, but he is convinced that these efforts will eventually pay off. We cannot take iPSCs as a black box, he urges. Only by thoroughly understanding how they work can we harness their power safely and responsibly.
