Since its emergence in humans 30 years ago, it has taken the lives of 39 million and infected another 35 million. Every year the US government spends close to 20 billion dollars globally to treat those infected, but a cure is still elusive. Instead, it continues to kill 6,000 people every day, making it the leading infectious killer in the world. The culprit is the human immunodeficiency virus, more commonly known as HIV.
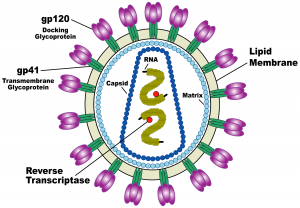
Why is HIV so deadly, and why have researchers still not found a cure for it? It turns out that HIV is an incredible biological machine that seems to evade and adapt to every defense mechanism our bodies have developed to defend us against bacteria and viruses. HIV resides in human blood, semen, vaginal fluid, and breast milk, and is transmitted from person to person through sexual activity, accidental events like the reuse of a used needle, and mother-to-child transmission. HIV cripples our immune systems by targeting T-helper cells, the centerpiece of the immune system. T-helper cells are so crucial because they identify the presence of foreign bodies and alert the immune system’s other components to attack the invaders. The collapse of this identification system results in Acquired Immune Deficiency Syndrome (AIDS) and impairs immune defense against a slew of other deadly conditions including cancer, tuberculosis, and additional viral infections.
HIV’s invasion strategy
HIV enters human cells in a brutal way. The virus has a membrane protein on its surface that latches onto a receptor protein on the T-helper cell’s surface, called a CD4 receptor. Then, using what’s called an HIV-1 Env spike protein, the virus drills into the membrane of the T-helper cell and uses a fishing-reel-like mechanism to pull the virus and T-helper cell membranes together, fusing the membranes and forcing open a pore. The virus then dumps its contents into the cell via this pore. The virus’s genetic sequence inserts itself into the cell’s DNA and takes charge, converting the cell into a virus-producing factory. The many viruses created in the host cell then leave to infect other cells.
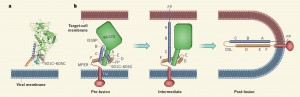
In almost all viral situations, the immune system targets and kills infected cells in order to stop the spread of the virus. But in the case of HIV, there’s a catch. The immune system can usually identify infected cells by the proteins they have on their surfaces. But an HIV-infected cell can stay dormant for many years and even decades, thereby evading detection by the immune system. Science has tried and so far failed to find a way to treat HIV once it has infested and becomes invisible inside normal-looking T-helper cells. For this reason, the key to combating HIV is to prevent the virus from entering the cell to begin with.
In order to do this, we need more knowledge about the mechanism the virus uses to fuse with T-helper cell membranes. As of one month ago, we didn’t know what this fusion machine looked like. But now we do.
Visualizing the mechanism
In October of this year, two breakthrough papers were simultaneously published in the journals Science and Nature—the world’s top two scientific journals—revealing for the first time the structure and dynamics of this fusion protein. This groundbreaking research was led by Dr. Walther Mothes, Associate Professor of Microbial Pathogenesis at Yale School of Medicine, Scott Blanchard of Cornell University, and Peter Kwong of the National Institutes of Health (NIH). The Science paper reveals the dynamics of the membrane (or “envelope”) protein on the surface of the virus, and the Nature paper displays its structure in the best resolution ever attained for the HIV-1 Env protein.
So how did they do it? To attain the structure, the group had to find a way to crystallize the envelope protein, which means they needed to isolate a homogenous array of the protein in a single uniform conformation. This was a monumental challenge, and Mothes’ work in conformational dynamics explained why.
It was already known that the envelope protein contains what are called the gp120 and gp41 subunits, two proteins each consisting of three components. Mothes reveals in the Science paper that the gp120 subunit constantly switches between one closed and two open conformations. It must open to fuse with the T-helper cell membrane, but Mothes also found that it can spontaneously open even in the absence of these cells. “If [the subunit] quickly adapts closed and open conformations, such a protein is impossible to crystallize. You never have a homogenous crystal,” Mothes says.
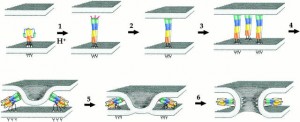
They visualized this conformational change with a technique called single molecule FRET (Fluorescence Resonance Energy Transfer). This technique differs from traditional population level FRET in that it targets single molecules instead of whole populations of molecules.
“The single molecule approach is important because when you look at how each molecule works, you see all the intermediates and steps that would be averaged out in a population,” Mothes explains. Mothes believes that the single molecule approach has the potential to dramatically speed up drug discovery, “basically cutting out ten years of the entire process of screening for inhibitors and understanding what they do.”
Single molecule FRET (smFRET) attaches fluorophores, which are dyes that emit light when excited by energy, to two different sections of the gp120 subunit so that changes in its conformation can be visualized by changes in fluorescence. One fluorophore emits energy in the form of light when excited by a certain wavelength of light energy, and this energy can pass onto the second fluorophore. But the energy can only be passed from one to the other if the two fluorophores are in close proximity, resulting in emission of light from the second fluorophore. If the fluorophores are far apart, energy cannot be transferred between them, and only the first fluorophore can emit light. By identifying which dye was emitting light, Mothes was able to see conformational changes in the gp120 trimer.
“Everybody thought [the trimer] was dynamic and moving, but they didn’t know that this machine opens up to adopt the final conformation where the receptor and coreceptor bind, all the time, spontaneously,” Mothes says. Now we know, and Mothes has seen it with his own eyes.
The next step: crystallization
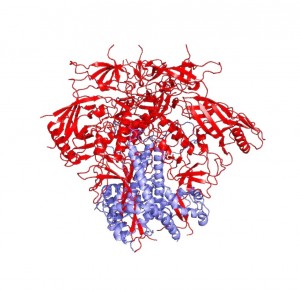
By identifying how the spike protein changes conformation, Mothes’ work explained what it took to crystallize the protein. In order to obtain a homogenous crystal, Peter Kwong’s group needed to use antibodies—proteins produced by the immune system—to bind to HIV-1 Env and keep it locked in one position. Normally, the immune system is able to easily produce antibodies that target envelope proteins like HIV-1 Env, but HIV has evolved to surround its envelope proteins with a glycan shield—a sugar coating. “It’s very clever,” says Mothes. Targeting sugars is not the immune system’s forte, and only very rarely does the immune system produce antibodies that can recognize patterns in the sugar coating or reach inside it to bind to the protein within. In rare cases, however, the immune system develops such effective antibodies. “What you find in some patients are antibodies that actually recognize the pattern of the glycan so that they recognize two or three glycans and a little piece of protein,” says Mothes.
Kwong used antibodies from people who had developed immunity to HIV and thus possessed these valuable antibodies in order to keep the envelope protein in one conformation and crystallize it. These antibodies are not only essential for protein crystallization, but are also potentially highly valuable for vaccine design.
One of HIV’s most problematic advantages is its extremely rapid mutation rate. Once the virus mutates its proteins, the antibodies the immune system has produced cease to be effective, and the body needs to make a new set of antibodies. Over time, however, the immune system begins to produce antibodies that recognize patterns that are common to all of the mutated forms of the protein. With more time and more exposure to different variants of the virus proteins, antibodies become effective against an increasing swath of variants. After a while, “the antibody that is made will also work for a hundred other HIV variants from different patients,” says Mothes. “So it will neutralize up to 90% of all HIV isolates we know.” Antibodies thus hold immensely significant clinical relevance.
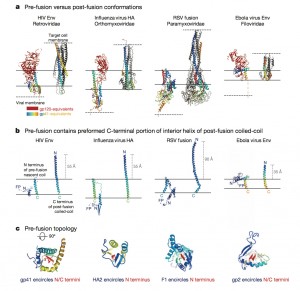
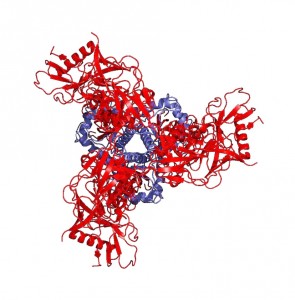
“Peter Kwong was able to use two of these antibodies, one of which could bind to gp41 and contribute to the stability of the gp41 subunit, helping to generate a crystal of the protein-antibody complex,” Mothes explains. He says that the significance of the structure lies in the resolution of the gp41 subunit, since the gp120 portion was resolved last December. The NIH group found that gp41 is a collection of helices and loops, and during fusion, the loops become straight helices, combining with the other helices to form one long helix that pulls the virus and T-helper cell membranes together for fusion. Mothes calls this pre-fusion conformation of loops and helices the “trigger-happy conformation,” given its energetic instability.
Knowing the structure of the gp41 subunit is truly a breakthrough in HIV research. “This is important,” says Mothes, “because the moment you can open Pymol [an online protein structure database] and look at the structure, you can use virtual screening software that can look at the pockets that a protein has and predict what small molecules may bind to these pockets.” And that’s only the tip of the iceberg. “Once we know where they bind, then you can make even more inhibitors and add something to the inhibitor that binds in an additional pocket to make it even more potent.”
Towards a vaccine
Visualizing the HIV fusion machine is not only pertinent to our knowledge of HIV. It turns out that other viruses including influenza and Ebola also use a very similar fusion machine to hijack cells, giving this discovery the potential to contribute to significant advances for the entire field of infectious diseases. The difference with HIV, however, is that HIV produces a chronic infection during which the virus escapes the immune system and adapts for a very long period of time. Ebola, on the other hand, is an acute infection that starts wreaking havoc immediately upon entering the human body. For this reason, Mothes believes that an Ebola vaccine will likely be available by next year.
Despite Mothes’ breakthrough visualization of the HIV fusion machine, there is still much progress to be made in the visualization and understanding of how HIV works. “We can look at the structure before and after, but everything in between is still just imagination,” Mothes says. Nevertheless, the two papers in Science and Nature mark a significant milestone in the history of HIV research and bring hope to field, since scientists can now work on developing vaccines that lock the HIV envelope protein in its inactive closed state. They are hoping that such vaccines will precipitate an immune response, giving the body enough time to develop immunity to the virus before the event of an infection. “All major HIV groups are coming out with trimers that are locked in this closed conformation,” says Mothes.
“Those are vaccine candidates. It’s happening.”

To read the author’s coverage of the rapid progress that has been made since this study, click here!
Extra Reading:
- Sanders, Rogier W., and John P. Moore. “A Stamp on the Envelope.” Nature 514 (2014): 437-38. 8 Oct. 2014. Web.
- Munro, James B., Jason Gorman, Xiaochu Ma, Zhou Zhou, James Arthos, Dennis R. Burton, Wayne C. Koff, Joel R. Courter, Amos B. Smith, III, Peter D. Kwong, Scott C. Blanchard, and Walther Mothes. “Conformational Dynamics of Single HIV-1 Envelope Trimers on the Surface of Native Virions.” Science 346.6210 (2014): 759-63. Web.
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto S, Gorman J, Huang J, Acharya P, Chuang Y, Ofek G, Stewart-Jones G, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, and Kwong PD. “Structure and Immune Recognition of Trimeric Prefusion HIV-1 Env.” Nature (2014) 514(7523): 455-61. Web.
- Video of HIV life cycle: https://www.hhmi.org/biointeractive/hiv-life-cycle
About the Author: Kevin Wang is a sophomore Molecular, Cellular, and Developmental Biology major in Ezra Stiles College. He is a Copy Editor for the Yale Scientific Magazine.
Acknowledgements: The author would like to thank Dr. Mothes for his time and enthusiasm. Dr. Mothes would like to recognize James Munro, a former member of the Mothes lab, for introducing the lab to many of the techniques used in this discovery.
