What constitutes a protein? At first, the answer seems simple to anyone with a background in basic biology. Amino acids, small organic compounds, link together into chains that fold into unique three-dimensional structures, which we call proteins.
Size matters in proteomics, or the scientific study of proteins. These molecules are typically complex, comprised of hundreds, if not thousands, of amino acids. A protein with demonstrated biological function usually contains no fewer than 300 amino acids. But findings from a recent study conducted at the Yale School of Medicine are challenging the notion that proteins need to be long chains in order to serve biological roles. That is, small size might not be an end-all for proteins.
The recent research was headed by the laboratory of Yale genetics professor Daniel DiMaio. First author Erin Heim, a PhD student in the lab, and her colleagues conducted a genetic screen to isolate a set of functional proteins with the most minimal set of amino acids ever described. The chains are short and simple, and yet they exert power over cell growth and the formation of tumors. Few scientists would have predicted that such simple molecules could have such huge implications for oncology, and for basic our understanding of proteins and amino acids.
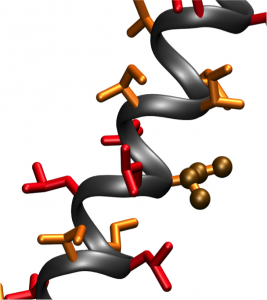
Engineering the world’s simplest proteins
There are 20 commonly cited amino acids, and their sequence in a chain determines the structure and function of the resulting protein. Most proteins consist of many different amino acids. In contrast, proteins identified in this study, named LIL proteins, were made up entirely of two amino acids: leucine and isoleucine.
Both of these amino acids are hydrophobic, meaning they fear water. The scientists at DiMaio’s lab were deliberately searching for hydrophobic qualities in proteins. An entirely hydrophobic protein is limited in where it can be located within the cell and what shapes it can assume. To maintain a safe distance from water, a hydrophobic protein would situate itself in the interior of a cell membrane, protected on both sides by equally water-fearing molecules called lipids. Moreover, the hydrophobic property reduces protein complexity by limiting the potential for interactions between the polar side chains of hydrophilic, or water-loving, amino acids. These polar side chains are prone to electron shuffling and other modifications, adding considerable complexity to the protein’s function and dynamics.
Heim and her group wanted to keep things simple — a protein that is completely hydrophobic is more predictable, and is thus easier to investigate as a research focus. “It’s rare that a protein is composed entirely of hydrophobic amino acids,” said Ross Federman, another PhD student in the DiMaio lab and another author on the recent paper.
The LIL proteins were rare and incredibly valuable. “[Using these proteins] takes away most of the complication by knowing where they are and what they look like,” Heim said. In terms of both chemical reactivity and amino acid composition, she said the LIL proteins truly are the simplest to be engineered to have a biological function.
Small proteins, big functions
What was the consequential biological function? Through their research, the scientists were able to link their tiny LIL proteins to cell growth, proliferation, and cancer.
The team started with a library of more than three million random LIL sequences and incorporated them into retroviruses, or viruses that infect by embedding their viral DNA into the host cell’s DNA. “We manipulate viruses to do our dirty work, essentially,” Heim said. “One or two viruses will get into every single cell, integrate into the cell’s DNA, and the cell will make that protein.”
As cells with embedded viral DNA started to produce different proteins, the researchers watched for biological functions. In the end, they found a total of 11 functional LIL proteins, all able to activate cell growth.
Of course this sounds like a good thing, but uncontrolled cell growth can cause a proliferation of cancerous cells and tumors. The LIL proteins in this study affected cell growth by interacting with the receptor for platelet-derived growth factor beta, or PDGFβ. This protein is involved in the processes of cell proliferation, maturation, and movement. When the PDGFβ receptor gene is mutated, the protein’s involvement in cell growth is derailed, resulting in uncontrolled replication and tumor formation. By activating the PDGFβ receptor, the LIL proteins in this study grant cells independence from growth factor, meaning they can multiply freely and can potentially transform into cancerous cells.
While this particular study engineered proteins that activated PDGFβ, Heim said that other work in the lab has turned similar proteins into inhibitors of the cancer-causing receptor. By finding proteins to block activation of PDGFβ, it may be possible to devise a new method against one origin of cancer. Even though the biological function in their most recent paper was malignant, Heim and her group are hopeful that these tiny LIL proteins can also be applied to solve problems in genetics.
Reevaluating perceptions of a protein
No other protein is known to exist with sequences as simple as those within the LIL molecules. Other mini-proteins have been discovered, but none on record have been documented to display biological activity. For example, Trp-cage was previously identified as the smallest mini-protein in existence, recognized for its ability to spontaneously fold into a globular structure. Experiments on this molecule have been designed to improve understanding of protein folding dynamics. While Trp-cage and similar mini-proteins serve an important purpose in research, they do not measure up to LIL proteins in biological functioning.
The recent study at the DiMaio lab pursued a question beyond basic, conceptual science: The team looked at the biological function of small proteins, not just their physical characteristics.
The discovery of LIL molecules and the role they can play has significant implications for the way scientists think about proteins. In proteomics, researchers do not usually expect to find proteins with extraordinarily short or simple sequences. For this reason, these sequences tend to be overlooked or ignored during genome scans. “This paper shows that both [short and simple proteins] might actually be really important, so when somebody is scanning the genome and cutting out all of those possibilities, they’re losing a lot,” Heim said.
Additionally, by limiting the amino acid diversity of these proteins, researchers were able to better understand the underlying mechanisms of amino acid variation. “If you want to gain insight into the heart of some mechanism, the more you can isolate variables, the better your results will be,” Federman said.
This is especially true for proteins. These molecules are highly complex, possessing different energetic stabilities, varying conformations, and the potential for substantial differences in amino acid sequence. By studying LIL proteins, researchers at the DiMaio lab were able to isolate the effects of specific amino acid changes at the molecular level. This is critical information for protein engineers, who tend to view most hydrophobic amino acids similarly. This study contradicted that notion: “Leucine and isoleucine have very distinct activities,” Heim said. “Even when two amino acids look alike, they can actually have very dissimilar biology.”
Another ongoing project at the lab involves screening preexisting cancer databases in search of short-sequence proteins. According to Heim, it is possible that scientists will eventually find naturally occurring cancers containing similar structures to the LIL proteins isolated in this study. This continuing study would further elucidate the cancer-causing potential of tiny LIL molecules.
To take their recent work to the next step, researchers in this group are looking to create proteins with functions that did not arise by evolution. The ability to build proteins with entirely new functions is an exciting and promising prospect. It presents an entirely new way of approaching protein research. The extent of insight into proteins is no longer bound by the trajectory of molecular evolution. Instead, scientific knowledge of proteins is being expanded daily in the hands of researchers like Heim and Federman.
About the Author: Emma Healy is a sophomore in Ezra Stiles college and a prospective Molecular, Cellular and Developmental Biology major.
Acknowledgements: The author would like to thank the staff at the DiMaio laboratory, with a special thanks to Erin Heim and Ross Federman for their time and enthusiasm.
Further Reading: Heim, E. N., Marston, J. L., Federman, R. S., Edwards, A. P., Karabadzhak, A. G., Petti, L. M., … & DiMaio, D. (2015). Biologically active LIL proteins built with minimal chemical diversity. Proceedings of the National Academy of Sciences, 112(34), E4717-E4725.
Cover Image: Art by Christina Zhang.