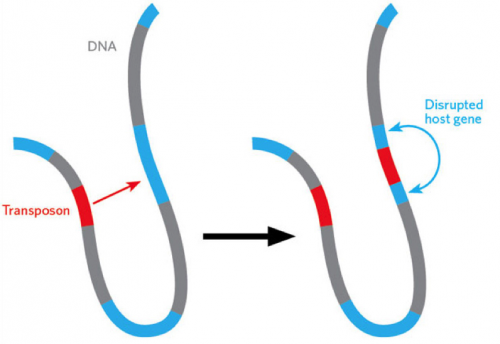
Imagine a gene being able to jump around within our genome. It would do so via a “cut and paste” mechanism, leaving a copy of the gene at its original site. These genetic tools have been termed “transposons” and are highly efficient in genome editing, making them viable for editing genes within the human genome. Dr. Tian Xu, Investigator of the Howard Hughes Medical Institute and Professor of Genetics at the Yale University School of Medicine, and his team have developed and studied a specific type of transposon complex, aptly named the piggyBac transposon, that can function in virtually all organisms and can easily “cut and paste” large amounts of DNA.
With increased efficiency and decreased expense, the transposon-based gene editing mechanism has become one of the most viable options for editing the human genome. A transposon complex consists of two main parts: the transposon itself and an enzyme. Transposons are DNA fragments that can move around the genome. The accompanying enzyme works in tandem with the DNA fragments, enabling the transposon to move, recognize, cut, and insert DNA in different locations.
In essence, this tool can be used for two key purposes, one of which is knocking down (“turning off”) singular genes. By comparing organisms with active genes and against those with silenced genes, scientists can study the function of these singular genes. In addition, transposon complexes have a transgenic function, meaning they can carry, transport, and insert sequences of DNA into new locations. However, this system has two major limitations. Although about 40% of human DNA consists of these transposon sequences, virtually all of them are inactive. Furthermore, most transposon-based complexes only function in their natural hosts.
After meticulous research, Xu and his team discovered that a transposon from moths called piggyBac could be modified and introduced into a mammal model. PiggyBac is the most active transposon. In a recent paper published in PNAS, Dr. Xu and his research associates studied the relationship between the piggyBac transposon and an enzyme complex called DNA-PK. DNA-PK was found to be integral to the function of this transposon complex, serving as a binding partner promoting the pairing of the DNA ends of the piggyBac transposon, therefore, helping the mobilization of the transposon. These findings have elucidated an evolutionarily conserved mechanism facilitating transposon activities in various organisms.
One of the most important findings in Dr. Xu’s study was that the newly discovered mechanism allows the design of a more efficient piggyBac transposon. It can be used to cut and insert large amounts of DNA, which has a variety of applications ranging from enabling researchers to easily study the effects of singular genes on different medical conditions to multiple uses in gene therapy. “We engineer the piggyBac transposon in the mouse genome, and therefore they can now jump in the mouse genome themselves…every new animal that has the transposon that is mobilized into a new location would have a mutated gene, Dr. Xu said. “You can have many animals with different mutated genes; now you can study function and therapeutic approaches.”