How Hi-P brings a new way to study protein-protein interactions
We use proteins for everything. Although they are most commonly known for fueling muscle growth, these large molecules are responsible for carrying out fundamental processes of life. Proteins can be antibodies that fight disease, enzymes that allow chemical reactions to take place in our body, hormones that lead to growth, and more.
In order to carry out their functions, proteins exhibit great structural diversity. For proteins, changing structure is the key to changing function. The protein’s shape determines the way it interacts with other molecules in the cell to serve its purpose. But even though we know a lot about what proteins are and what they do, we still lack knowledge about how they interact with other molecules.
Before, scientists could only investigate proteins individually, one interaction at a time. But last summer, Jesse Rinehart, Professor of Cellular and Molecular Physiology at Yale, and his lab successfully developed a way to observe all the interactions taking place within a cell, also known as the proteome scale. The Rinehart lab’s method can be broken down into two general steps: (1) building a protein library and (2) using Hi-P, a readout method they invented, to analyze the protein interactions.
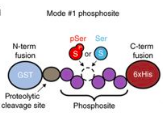
A protein library is a collection of a wide variety of human proteins generated by bacteria. To get them to produce the human (instead of bacterial) proteins, segments of DNA called plasmids are inserted into the bacteria. The plasmids designed by the Rinehart lab, however, are special because they can manipulate the bacteria to make either a phosphorylated or non-phosphorylated protein. Before, phosphorylated proteins could not be produced. Phosphorylation modifies the protein by adding a chemical group, phosphate, to its structure. This process not only allows the bacteria to produce the exact same proteins that we make, but also gives the protein a tag that allows it to be identified later.
Once this protein library is produced, it needs to be interpreted, and that’s where Hi-P comes in. “Hi-P is a readout for if a protein-protein interaction occurs,” Rinehart said. “It allows us to find how our library interacts with other types of protein.” As the phosphorylated proteins produced earlier bind with other human proteins, researchers can sequence the DNA of the phosphorylated protein to identify it, allowing scientists to retroactively connect the interaction to the protein.
“That’s a big deal because the information tells us directly what’s going on at the protein level,” Rinehart said.
As of proof of concept, the Rinehart lab applied their method on interactions involving 14-3-3 and WW2 proteins, which have already been well-studied. Now, the next steps are to expand the technology and look at uncharacterized protein interactions. “We build the technology and do the confirmatory work first. Now we can unleash it and do more exploratory work,” Rinehart said.
Rinehart hopes that this is just the first step to major discoveries down the line. Protein interactions dictated by phosphorylation lie at the heart of cancer and diabetes, so studying these interactions may help us understand and further tame these diseases.