Image courtesy of Yang Liu.
To understand the function of a cell, we must first understand the role of genes. Think about a section of tissue as small as a single hair follicle: the activity of tens of thousands of genes forms a blueprint for this fraction of space—packed full of organelles, cellular fluids, and more. How can we get a complete view of this complex space and its inner workings?
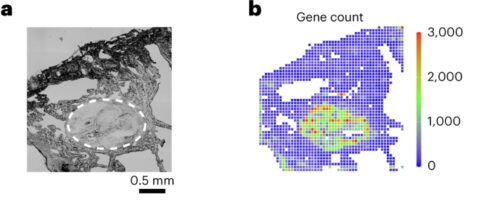
Yang Liu, an assistant professor at Yale University, sought to visually describe these relationships through a method called spatial CITE-sequencing, where transcriptomic data can be associated with sections of tissue via a series of protein tags, antibodies, and short RNA sequences. Researchers then form a 2D map and pinpoint the locations of hundreds of genes simultaneously. Previously, CITE-sequencing could only sequence a limited number of targets with low efficiency. Now, the technique can sequence hundreds of genes simultaneously. This technology will help grow global databases for RNA coding sequences, which are valuable for understanding the distinct roles of genes and generating possible treatments for genetic diseases.
When working with a process as novel as CITE-sequencing, a technique that has only been available for five years, the successes are rewarding, but the failures are frequent. “Whenever you solve [one problem] there is another. But incremental progress is still progress, so taking a big problem and breaking it up into smaller ones is essential,” Liu said. For example, to solve an issue of keeping thin fragments of tissue in place for experimentation, the team has been developing prototypes of a specialized tissue clamp. Spatial transcriptomics is expanding quickly, and this Yale research team is leading the charge to add to our knowledge of gene activity. An increase in funding for the field has promoted protocol optimization, decreased experimental costs, and improved accessibility for CITE-sequencing technology.