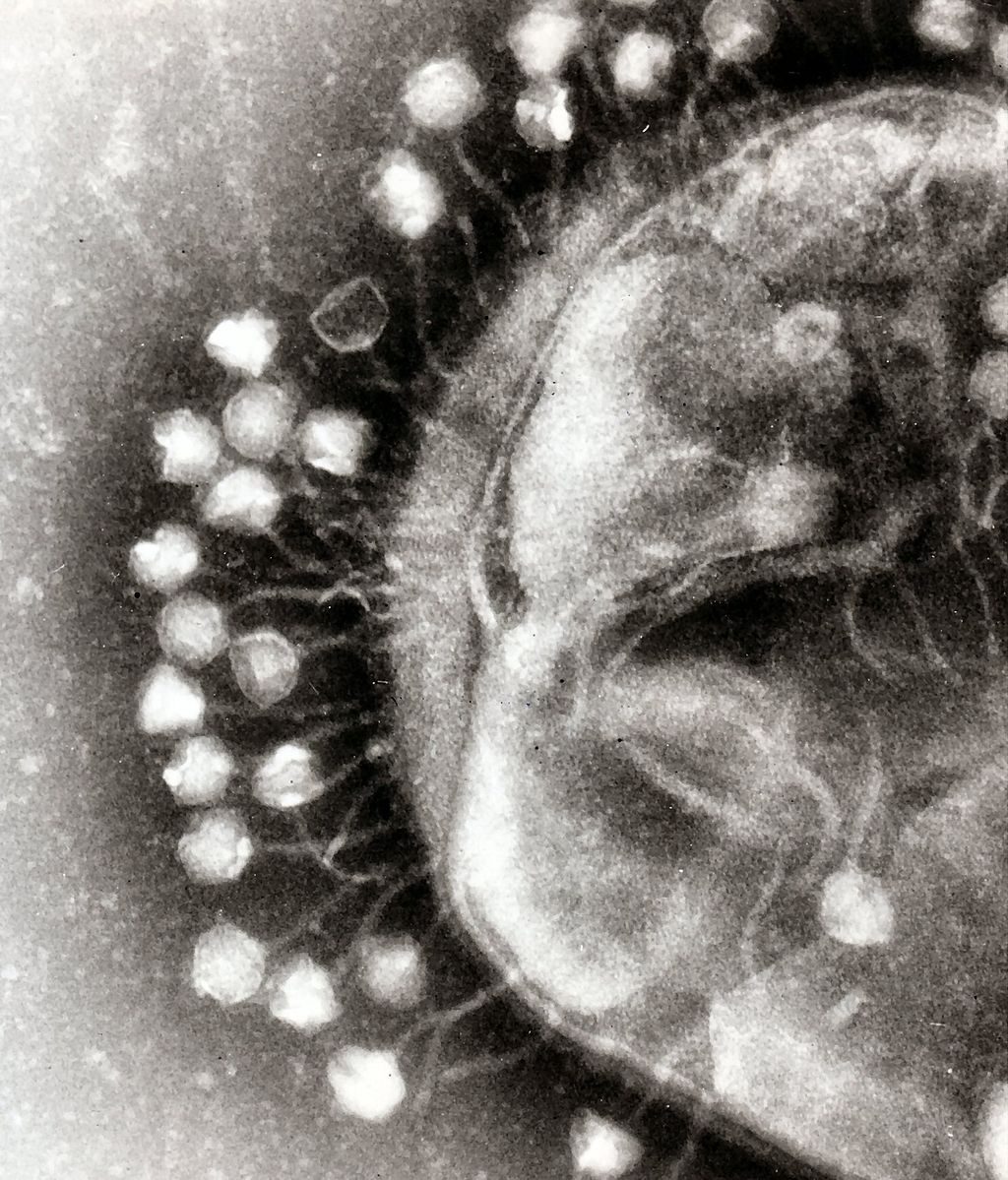
In 2020, the World Health Organization labeled antimicrobial resistance as one of the greatest threats to global health. Resistance occurs when bacteria evolve defense mechanisms that counter the drugs designed to destroy them.
Katie Kortright and her colleagues in the Yale Department of Ecology and Evolutionary Biology are leading the search to find a worthy opponent for antimicrobial resistance. For decades, scientific literature has pointed to the bacteriophage, or phage for short, as a possible solution. A phage is a viral parasite that infects a bacterial host. Translating appropriately to “devour” in Greek, the phage reproduces by bursting out of its host, killing the bacteria.
In nature, when two species interact, coevolution can occur, where organisms genetically adapt to each other over generations. Two forms of coevolution may happen under different conditions: arms-race (ARD) and fluctuating selection (FSD) dynamics. ARD is a back-and-forth acquisition of attack and defense mechanisms while FSD implies a continuous cycle of mutual resistance. Tackling antimicrobial resistance, Kortright aimed to find out which form, if any, would exist in bacteria-phage interactions.
To accomplish this goal, Kortright exposed colonies of bacteria to phages for ten days. She investigated coevolution by sequencing the genome of the bacteria, and from the genome, she found that coevolution occurred through both ARD and FSD dynamics. This finding demonstrates that different mechanisms of coevolution can occur under a single bacteria-phage system. The reason for this phenomenon, however, remains unknown.
The battle is far from over. Before phage therapy becomes a feasible combatant against antimicrobial resistance, it is imperative to identify every evolutionary consequence. “We are never going to run out of things to study,” Kortright said.